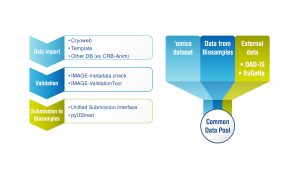
Discover the portal that integrates and presents data from gene banks with genomics data and geographical information systems data.
One of the key challenges facing the management and conservation of animal genetic resources is the integration and availability of the vast amount of information that is stored within more than 60 animal gene banks spanning across 20 European countries. In order to provide access to the huge amount of heterogenous data distributed across gene banks in different locations, storage formats and language, a new concept was needed.
Addressing this challenge was one aim of the H2020 Innovative Management of Animal Resources project (IMAGE). The IMAGE’s web portal integrates data from gene banks and collections, with genomics data, geographical information systems data, and other information generated by the IMAGE project.
The solution implemented is comprised of the following four factors:
- A well-defined metadata rule set ensuring high quality and comparable data across the diverse collections originating in different storage formats and languages;
- The development of a single Inject-tool helping gene bank managers to enhance, standardise, tag and submit their gene bank data to a Common Data Pool that integrates all gene bank records from across Europe;
- The sustainability offered by archiving of these data within the EBI BioSamples public archive; and
- A bespoke data portal, integrating gene bank metadata with generated ‘omic’ datasets from within IMAGE and cross referencing to other gene banks and breeding database resources, such as those hosted by the Food and Agriculture Organization (FAO).
Building a BioSamples resource for IMAGE
The Inject tool is a web tool which curates data by validating against the IMAGE metadata ruleset to guarantee standardised and high-quality data. The tool further assists users by handling the brokering of data to BioSamples and automating the majority of the submission steps. BioSamples assigns a universally recognised unique identifier to each organism and specimen. Minimum identification includes species, breed, sex, and organism part.
All IMAGE data archived with BioSamples is also made available to the community through the IMAGE data portal that collates all of the IMAGE samples and genomic data into a single interface. The aim is to provide a user-friendly interface for accessing breed related (meta) data of germplasm/DNA collections and related genomic data. It provides easy access to metadata on breed specific genetic collections and related databases at European level through a web portal, and to allow/facilitate connection between databases through appropriate interfaces
The IMAGE portal is closely connected to the European Gene Bank Network portal – a network of officially recognised national gene banks for long term conservation, managed by the European Regional Focal Point for Animal Genetic Resources (ERFP). One of the objectives of EUGENA is to enable access to the register of EUGENA gene banks in Europe. Gene banks covered by EUGENA will be a subset of all existing genetic collections in Europe, as it only covers officially recognised gene banks for long term conservation.
The portal is currently displaying information uploaded for three countries for more than 32,000 samples and is under continuous improvement. Predictive text-based search across all IMAGE metadata fields is used and it also allows the downloading of selected information in order for it to be analysed using the diversity browser. Uploading of samples, facilitated by the Inject Tool, is still undergoing for most countries. Once completed, it will represent the most comprehensive collection of gene bank information at a European level and beyond.
Exploiting the data
Within the portal, a Geographic Information System tool is included in order to assist the user in identifying/storing the geographical origin of the samples, as well as displaying individual/population genetic parameters and biological attributes through interactive maps. Querying across all types of data is also expected to facilitate targeted searches to identify genetic material of interests residing somewhere in the partner gene banks and collections. Furthermore, starting from data derived from the portal, computing tools and methods have been developed to browse the diversity of sample and/or genomic data.
In addition to this, an interactive web interface to guide the use of genetic material was created. It allows selective downloading of collection and genotype information to be leveraged using a linked R software package called ‘MoBPS’. This Modulary Breeding Program Simulator provides a computationally efficient and flexible framework to simulate complex breeding programs or conservation programs and compare their economic and genetic impact. The primary design philosophy behind MoBPS is to provide the community with a tool that is able to simulate all breeding programs.
To conclude, the portal presents search tools and summary statistics and links to the analysis tools, as well as to the DAD-IS and EUGENA databases. Detailed documentation is available on the development of the portal, the common data pool, the Inject tool and the diversity browser, genotype data submission guidelines and the import script. This information is available on the github website.
Michèle Tixier-Boichard
IMAGE coordinator
INRAE
Alessandra Stella
WP leader in charge of the data portal
CNR
michele.boichard@inrae.fr
alessandra.stella@ibba.cnr.it
www.imageh2020.eu
Please note, this article will also appear in the second edition of our new quarterly publication.