The evolution of antibiotic resistance and combatting the challenges posed by multidrug-resistant bacteria.
Alexander Fleming was awarded with Howard Florey and Ernst Boris Chain the Nobel Prize in 1945 for the discovery of Penicillin, an antibiotic which revolutionised the world. However, resistance was established very early and Fleming reported this observation in his Nobel speech:
“…But I would like to sound one note of warning. Penicillin is to all intents and purposes non-poisonous so there is no need to worry about giving an overdose and poisoning the patient. There may be a danger, though, in underdosage. It is not difficult to make microbes resistant to penicillin in the laboratory by exposing them to concentrations not sufficient to kill them, and the same thing has occasionally happened in the body”.
Evolutional history of antibiotic resistance and the emergence of multidrug-resistant bacteria
Antibiotic resistance is a natural phenomenon and an evolutionary survival process of bacteria; though it has been widely distributed throughout mankind (Pawlowski et al., 2016), its development and dissemination have become accelerated by the overuse or misuse of antibiotics last few decades. The selective pressure exerted by antibiotics has dramatically increased drug-resistant bacteria (pathogens and commensals) (Fair and Tor, 2014) and thus contributed to the World Health Organization (WHO) announcing it as a global threat to health and economy. Centers for Disease Control and Prevention (CDC) estimates in the United States, at least 2 million people annually get an antibiotic-resistant infection, and at least 23.000 people die. The health burden of infection with antibiotic-resistant bacteria is high in Europe with an estimated 671.689 infections in 2015, of which 63.5% were associated with healthcare (Cassini et al., 2019). Furthermore, the global annual cost of antibiotic resistance could increase to 10 million deaths and $100 trillion by 2050 (O´Neill, 2016). Fighting this threat is a public health priority that requires a collaborative global approach across sectors.
Bacteria predominantly evolve to resist antibiotics through mutation and horizontal gene transfer (HGT). Spread of antibiotic resistance elements was enhanced by HGT mobilizing antibiotic resistance genes (conjugation and transduction) and allowing pathogenic bacteria to acquire resistance mechanisms from commensal bacteria. Transfer of antibiotic resistance genes occurs between species and even genera taxonomically not related, which leads to the overwhelming superbugs. This compensatory evolution of bacteria may alleviate the fitness cost and therefore drives to the spread of antibiotic resistance throughout different ecosystems.
Animal products as main reservoir of multidrug-resistant bacteria
Microbial infections caused by contaminated food supplies frequently remind us of the complex food-web that links us with animal, plant, and microbial populations globally. The presence of high microbial loads, especially pathogens in animal products destined for human consumption, constitute a challenge and reinforce the importance of controlling zoonotic pathogens in meat and other animal products through a complete and continuous farm-to-fork examination. Animals commonly harbour bacteria in their skin, hair and wool, noses, urogenital and oral cavities. As well as this, bacteria from intestinal tracts can spread from the animals to the environment, and from the slaughterhouse to the food chain. (Sobsey et al., 2006). Furthermore, slaughterhouses pose a great threat in harbouring antibiotic resistant bacteria, due to the use of metaphylactic and prophylactic antibiotics (previously used for growth promotion, but now banned in Europe since 2006) in veterinary, clinical therapy and in agriculture (Food and Agriculture Organization of the United Nations [FAO], 2015, 2016). In this context, new food-borne pathogens often emerge from animal reservoirs. According to the European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018 (EFSA, 2020), overall multiple drug resistance (MDR) was increased in zoonotic and indicator bacteria from humans, animals and food. Lives are at risk, thus measures to prevent food poisoning and curtail behavior that promote antibiotic resistance are urgently needed.
Strategies developed to reduce and stop the spread of antibiotic resistance
To face the increasing and multifaceted problem of antibiotic resistant pathogens (superbugs) in meat food chain and in the environment (Economou and Gousia, 2015), multiple strategies have been developed, as the incidence of environmental contamination varies greatly between countries and regions. In this sense, management strategies and preventive medicine consisting of reduction of antibiotic use in animals, has increased focus to hygienic practices, feed quality and environmental conditions, and the use of alternative growth promoters were proposed to reduce diseases in animals and subsequently, use of antibiotics. However, reduction of antibiotic use could have minimal effects on the preestablished antibiotic resistant bacteria which developed a compensatory evolution in response to antibiotic pressure.
As a shared global challenge, the use of disinfection strategies was widely extended to reduce pathogens, however the consequences on antibiotic resistance gene spread were poorly understood. Most users of disinfectants would not expect that biocidal agents may cause antibiotic resistance themselves. For example, triclosan was used for decades in antimicrobial soaps in the USA and considered to be safe and effective; however in 2016, the US Food and Drug Administration banned 19 active ingredients, including triclosan for antimicrobial soaps domestically used by the general population (Department of Health and Human Services, 2016).
Due to triclosan’s associated risks to the spread of antibiotic resistance throughout the environment, other biocidal agents are being used for disinfection in healthcare, veterinary medicine, food production, food handling or in domestic settings; they may also carry risks of enhancing antibiotic resistance, especially during low-level exposure (Kampf, 2018a). Sublethal concentrations of biocidal agents may cause antibiotic resistance by promoting an adaptive response in bacteria (Kampf, 2018b). On the other hand, biofilm formation confers increased antimicrobial resistance through a variety of mechanisms facilitated by an intracellular communication mechanism known as quorum sensing, thus biofilms are 10- to 1000-fold less susceptible to antimicrobial agents than the planktonic form of the same bacterium (Sereno et al., 2017). In poultry processing plants, the use of sub-optimal concentrations of a commonly applied biocide (quaternary ammonium, organic acids, chlorine, alkaline detergents) has facilitated the persistence of Salmonella biofilms and the mechanisms that increase their tolerance to disinfectants and cross-resistance to antibiotics (Cadena et al., 2019). Recently, Bridier et al. (2019) showed that cleaning and disinfection during a short time period did not lead to the selection of Salmonella with higher resistance toward antibiotics and biocides, but markedly impacted the composition of bacterial populations associated to slaughterhouse surfaces.
The presence of mobile genes represents a great challenge and could result in a pandemic with serious health and socio-economics repercussions (Founou et al., 2016), with food trade being a contributing factor because of the spread of these genes to humans via meat products. Slaughterhouses and meat processing represent a nexus of the “One Health” (or “One Medicine”) initiative that links human, animal and environmental health. Therefore, control of antibiotic resistance genes spread by adequate disinfection procedures in slaughterhouse becomes necessary to protect the health of human and animals.
Despite different disinfection approaches applied in different industries, food contamination and bacterial infections with emergent and antibiotic resistant pathogens are growing, causing a serious global health problem. The emergence of antibiotic resistant bacterial strains has been dramatically increasing, which has made antibiotic choices for infection control increasingly limited and more expensive. Thus, alternatives for antibiotic therapy and other strategies to reduce selection and spread of antibiotic resistance is a key priority.
Adequate disinfection strategy to avoid/reduce antibiotic resistant bacteria spread
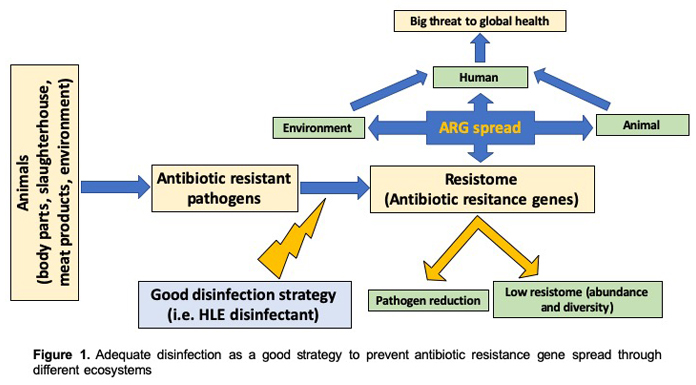
A good disinfection strategy based on compounds able to efficiently eradicate multiple species of bacterial pathogens and are also friendly to the environment, is the key measure to counter the spread of potential pathogens in different ecosystems. This approach may reduce the emergence of new antibiotic resistant pathogens regardless of the selective pressure of antimicrobials used (antibiotics or biocides) (Figure 1). The efficacy of a new disinfectant product, HLE, has emerged, and inhibits multiple species of planktonic and biofilm bacterial cultures. (Abriouel et al., 2018). The HLE disinfectant comprised EDTA, lactic acid and hydrogen peroxide, and our data indicated that the disinfectant had effective antimicrobial and anti-biofilm activity, and was able to destabilize biofilm structures even at low concentrations. The synergistic effect of EDTA and both antimicrobials (lactic acid and hydrogen peroxide) allowed pathogen eradication and was also able to inhibit the expression of multidrug efflux pumps implicated in resistance. This strategy allowed elimination of pathogens and could stop the emergence and the spread of antibiotic resistant bacteria in the food chain and the environment by decreasing efflux pump expression, thus increasing the fitness cost of bacteria. Therefore, the antibiotic resistance genes present in both pathogenic and non-pathogenic bacteria will be gradually reduced.
Biofilms are complex structures adopted by bacteria as a good survival strategy in different environments, thus they become a great challenge in food industries and medical settings, due to their high resistance to antimicrobials. Alternatives to antibiotics or biocides will be based on biofilm-preventing and disrupting enzymes, which guarantee the success of antimicrobial efficacy. HLE disinfectant could be a good alternative as a biofilm-preventing strategy and as a biofilm-disrupting structure because it is able to produce a greater porous structure with larger channels among the cell clusters versus the control (Abriouel et al., 2018).
The consequences of antibiotic resistance maybe aggravated by the current SARS-CoV-2 pandemic, as antimicrobial resistance crisis is considered a pandemic hidden threat. Furthermore, antibiotics have been widely used to eliminate possible bacterial superinfections in COVID-19 patients and thus increased level of antibiotic resistant bacteria was detected.
References
Abriouel, H., Lavilla Lerma, L., Pérez Montoro, B., Alonso, E., Knapp, C.W., Caballero Gómez, N., Gálvez, A., Benomar, N. 2018. Efficacy of “HLE”—a multidrug efflux-pump inhibitor—as a disinfectant against surface bacteria. Environmental Research 165: 133–139.
Bridier, A., Le Grandois, P., Moreau, M.H., Prénom, C., Le Roux, A., Feurer, C., Soumet, C. 2019. Impact of cleaning and disinfection procedures on microbial ecology and Salmonella antimicrobial resistance in a pig slaughterhouse. Scientific Reports 9: 12947.
Cadena, M., Kelman, T., Marco, M.L., Pitesky, M. 2019. Understanding Antimicrobial Resistance (AMR) Profiles of Salmonella Biofilm and Planktonic Bacteria Challenged with Disinfectants Commonly Used During Poultry Processing. Foods 8: 275.
Cassini, A., Hogberg, L.D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G.S., Colomb-Cotinat, M., Kretzschmar, M.E., Devleesschauwer, B., Cecchini, M., et al. 2019. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 19(1): 56–66.
Economou, V., Gousia, P. 2015. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 8: 49–61.
European Food Safety Authority and European Centre for Disease Prevention and Control. 2020. European Food Safety Authority and European Centre for Disease Prevention and Control. EFSA J. 18(3): 6007.
Fair, R.J., Tor, Y. 2014. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 6: 25–64.
Founou, L.L., Founou, R.C., Essack, S.Y. 2016. Antibiotic resistance in the food chain: a developing country-perspective. Front. Microbiol. 7: 1881.
Food and Agriculture Organization of the United Nations [FAO]. 2015. Status Report on Antimicrobial Resistance. Rome: Food and Agriculture Organization of the United Nations.
Food and Agriculture Organization of the United Nations [FAO]. 2016. The FAO Action Plan on Antimicrobial Resistance 2016-2020. Rome: Food and Agriculture Organization of the United Nations.
Kampf, G., Introduction. In: Kampf G., editor. Antiseptic Stewardship: Biocide Resistance and Clinical Implications. Springer International Publishing; Cham, Switzerland: 2018a. pp.1-7.
Kampf, G. 2018b. Biocidal Agents Used for Disinfection Can Enhance Antibiotic Resistance in Gram-Negative Species. Antibiotics (Basel). 7(4): 110.
J. O’Neill, “Tackling drug-resistant infections globally: Final report and recommendations,” The Review on Antimicrobial Resistance (2016).
Pawlowski, A.C., Wang, W., Koteva, K., Barton, H. A., McArthur, A.G., Wright, G.D. 2016. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 7: 13803.
Sereno, M.; Ziech, R.; Druziani, J.; Pereira, J.; Bersot, L. 2017. Antimicrobial Susceptibility and Biofilm Production by Salmonella sp. Strains Isolated from Frozen Poultry Carcasses. Braz. J. Poult. Sci. 19: 103–108.
Sobsey, M.D., Khatib, L.A., Hill, V.R., Alocilja, E., Pillai, S. 2006. “Air quality and emissions from livestock and poultry production/waste management systems,” in Animal Agriculture and the Environment: National Center for Manure and Animal Waste Management White Papers, eds J. M. Rice, D. F. Caldwell, and F. J. Humenik (USA: ASABE publication), 609-666.