Marineholmen RASLab’s CEO Dr Mark Powell and Research Coordinator Dr Irene Roalkvam explain what genome sequencing can tell us about biofilter bacteria in recirculating aquaculture systems.
Recirculating aquaculture systems (RAS) operating at a high degree of water recirculation depends on an array of water purification technologies to maintain good water quality for large fish stocks. An active biofilter with high capacity of microbial nitrification is vital for a successful removal of ammonia. However, detailed knowledge on microbial colonisation, development, and influencing factors on these bacteria is still deficient today, despite the use of biofilter technology in aquaculture for decades.

Biofilter diversity at Marineholmen RASLab
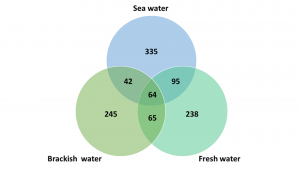
At Marineholmen RASLab, the biofilters were originally inoculated with commercial enrichment culture in May 2020, but have developed to be specific biofilters adapted to three different salinities over the last two years: fresh water (0ppt NaCl), brackish water (15ppt NaCl), and sea water (34ppt NaCl) (Fig. 1). To obtain detailed information about the nitrifying bacteria in biofilms from all three types of biofilter, targeted 16S rRNA genes from the entre microbial communities were amplified and sequenced using Illumina sequencing technology.
After two years of sustaining biofilters in the three storage tanks, the sequencing results revealed significant differences in microbial communities present in the specific types of biofilter. Overall, the diversity in the biofilter for brackish water was much lower compared to the biofilters for fresh water and sea water. Furthermore, the number of common bacterial taxa (bacteria present in all three biofilter types) was only 64, while the number of unique bacterial taxa were 238-335 (Fig. 2).
This confirms that the microbial communities in the biofilters are affected by environmental factors, such as different salinity, and that various bacterial species will adapt to the different conditions and thereby form different biofilters.
Within the complex microbial community in biofilters, the nitrifying bacteria usually establish slowly over time, and the colonisation period of new biofilters can vary from weeks to several months. In general, the ammonia oxidising step in nitrification is, in most cases, established before the nitrite oxidising step, and the nitrification process in RAS appear to be associated with a small group of bacteria, including the ammonia oxidisers Nitrosomonas, Nitrosospira, and Nitrosococcus; and the nitrite oxidiser Nitrospira.
The brackish water biofilter
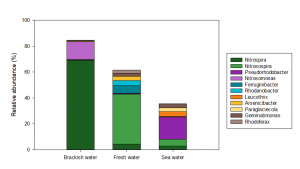
The brackish water biofilter at Marineholmen RASLab is considered robust and efficient, with high tolerance for changes in water quality or operational conditions and with a relatively short lag phase before resuming normal activity levels after RAS start-up. This biofilter was dominated by two different species of Nitrospira (69.1% relative abundance, of which N. marina had 66.2%), but also comprised two different species of Nitrosomonas (13.6% and 0.32% relative abundance) (Fig. 3).
Fresh water and sea water biofilter
The fresh water and sea water biofilters are generally less robust, as they have a considerably longer lag phase before reaching adequate activity level in the biofilters after RAS start-up, and they also seem to initiate a lag phase or periods with a decrease in activity level under changing operational conditions. As expected, these biofilters contained different nitrifying bacteria (Fig. 3). Both biofilters had much lower relative abundances of Nitrospira compared to the brackish water biofilter, and other species than N. marina comprised the highest relative abundance in both biofilters.
Furthermore, the fresh water biofilter was dominated by the genus Nitrosospira (38.5% relative abundance), which was also present at high relative abundance in the sea water biofilter (5.0% relative abundance), but most likely associated with different species. The fresh water and sea water biofilters also comprised small amounts of Nitrosomonas (<0.6% relative abundance), which were also represented by different species than in the brackish water biofilter.
Recirculating aquaculture systems analysis
Bacteria involved in both steps of nitrification were detected in all biofilters at Marineholmen RASLab. However, the biofilters clearly did not develop equally as different salinities enriched different nitrifying species, having different growth requirements or responses to changing conditions, even though they still served similar functions in the biofilters. Illumina sequencing of the biofilter biofilm is a powerful tool to examine the entire microbial community structures at genus and species level.
Therefore, understanding how the microbial community changes as conditions change under aquaculture production demands and operations can give an early warning to risk of microbial community shifts that may lead to catastrophic failure of the biofilter and thereby also the production outcome. Yet, next-generation sequencing is another approach RASLab is using in our drive for “innovating the future of aquaculture.”
Please note, this article will also appear in the tenth edition of our quarterly publication





